Close, but no C(igar) -- Optimising Numerics-Heavy Python Code with Numba
3th October 2022Introduction
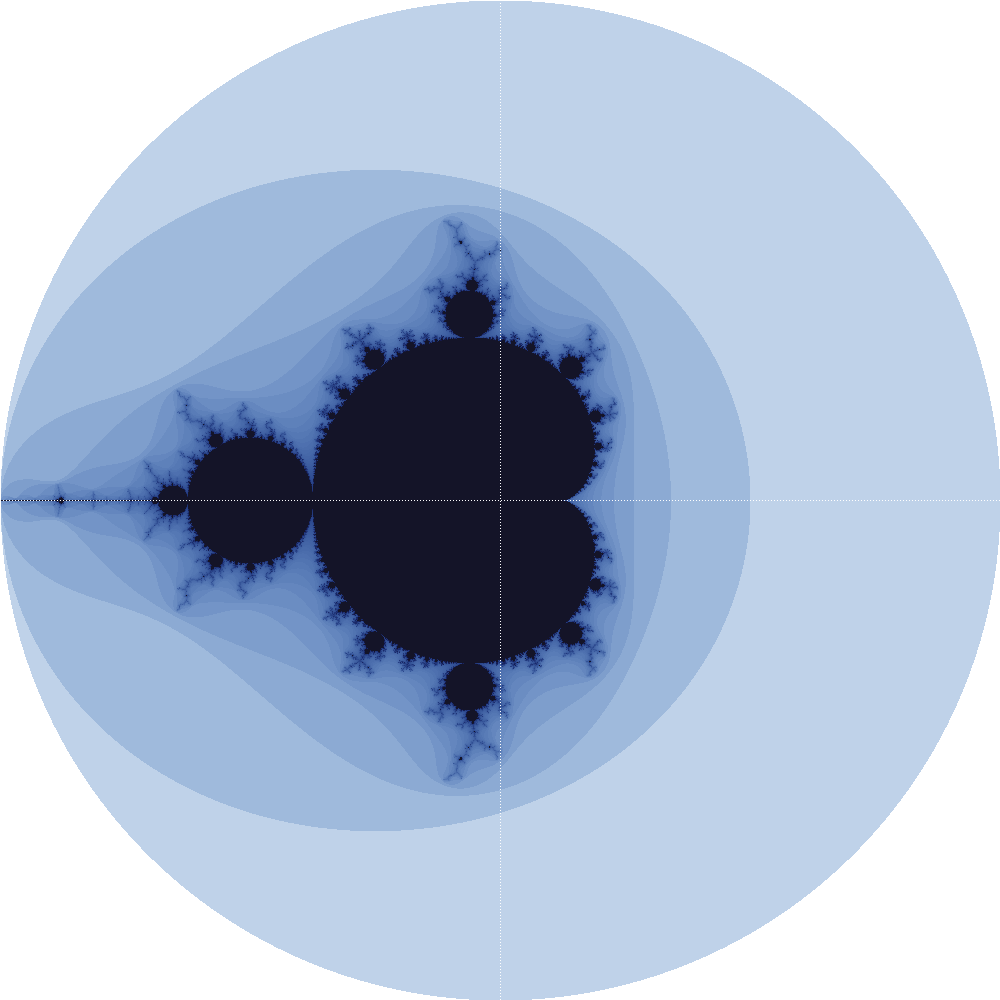
We all know that C is fast and Python is slow -- at least to first order ;-). I recently implemented a renderer for the famous Mandelbrot Set in plain C (see here for the source code), and as always, I was pleasantly surprised by the speed: rendering 10M pixels (yielding a PNG file of approximately 380 MiB) took only slightly longer than a minute on my pretty old machine from 2012. See here for an exemplary (downscaled) output with only 100k pixels, which took approximately 700ms to compute and render:
Comparing it to Python
A first comparison with an implementation written in 'naive' 'list-of-lists'-style Python was underwhelming. Based on mean and standard deviation of the execution times of three replicates, C definitely outperforms Python approximately 40-fold (see here for all the raw data). Just compare the first two bars in the following plot:
Using Python's integrated complex datatype (see here for the source code) significantly reduced computation time, however the effect is rather small (see third bar in the plot above, showing a reduction of required time by approximately 15%).
If we make use of numpy arrays for storing our computed values instead of using a naive 'list of list' approach as before (see here and here for the respective implementations), computation times do not change significantly (see bars 4 and 5). This does not come as a surprise, since the majority of time can be expected to be spent on computing, instead of storing values in memory.
The numba library
To further increase performance, we may use the numba Python library, which "translates Python functions to optimized machine code at runtime using the industry-standard LLVM compiler library." It is claimed that "Numba-compiled numerical algorithms in Python can approach the speeds of C or FORTRAN."
Bar 6 shows the performance of my Python implementation using numba. It should be remarked that (at least in this case) using numba literally boils down to importing the library and adding one single decorator directly in front of the "critical" routine as follows:
@numba.njit # <---- Magic happens here!
def iterate(c_r, c_i):
zn_r = X0_R
zn_i = X0_I
i = 0
while i < MAXITER:
zn_r_2 = zn_r * zn_r
zn_i_2 = zn_i * zn_i
if zn_r_2 + zn_i_2 > LIMIT_2:
break
zn_i = 2 * zn_r * zn_i - c_i
zn_r = zn_r_2 - zn_i_2 + c_r
i = i + 1
return iThis single line has reduced computation time by approximately 95% alone! However, it still significantly exceeds the computation time of my implementation in C, requiring approximately twice the time.
Very interestingly, using Python's complex datatype actually significantly decreased performance by approximately 66% (see the last bar in the plot above). I assume, numba is not smart enough to keep down the number of arithmetic operations required for the Mandelbrot iteration. My "manual" way (see above) is rather optimised, as it avoids calculating squares twice in the inner loop, using them both for calculating the absolute value of \(z\), and for calculating the next iteration step.
Some more remarks
I also tried the fastmath option provided by numba (see here), which decreases floating point accuracy in order to get faster computation, but this did not significantly altered computational performance.
I also tried this clever approach based on numpy matrix operations (see here for my implementation), however performance actually decreases, likely because all the slicing and dicing of arrays costs more time than numpy vectorisation yields.
Conclusions
First, nubma makes stuff extremely fast. While it can (at least in this example) not reach the performance of C, it offers a tremendous "return on investment" time-wise, as it generally boils down to adding a decorator to critical functions in an existing Python codebase. Thus, there's no need to rewrite routines from scratch.
Second, there is still some room for optimisation, as I have not used numba's feature for parallelisation, since I aimed for a fair comparison with my single-cored C implementation. For now, I assume setting up parallel mode is likely easier done in Python using numba then in plain C. Thus, the potential "return on investment" might be even larger.
And finally, some manual optimisation can still yield significant performance gains, especially if they concern the choice of efficient algorithmic approaches (see my remark above regarding the complex datatype) and/or the choice of efficient data structures. This, of course, requires that the programmer has some "domain knowledge" about the problem he is working on (in our example, it boiled down to the simple mathematical fact that we can re-use some computed values instead of recomputing them in the inner loop), so that he is able to see some optimisations which are not immediately obvious to a software library.